
CRAC is the only RNA-Seq mapping software that include the discovery of transcriptomic and genomic variants like splice junction, chimeric junction, SNVs, Indels in a single analysis step using a built-in error detection method enabling high precison and sensitivity.
Find more about CRAC's concept here.
What's new?
A pipeline to analyse non-coding variants which uses CRAC
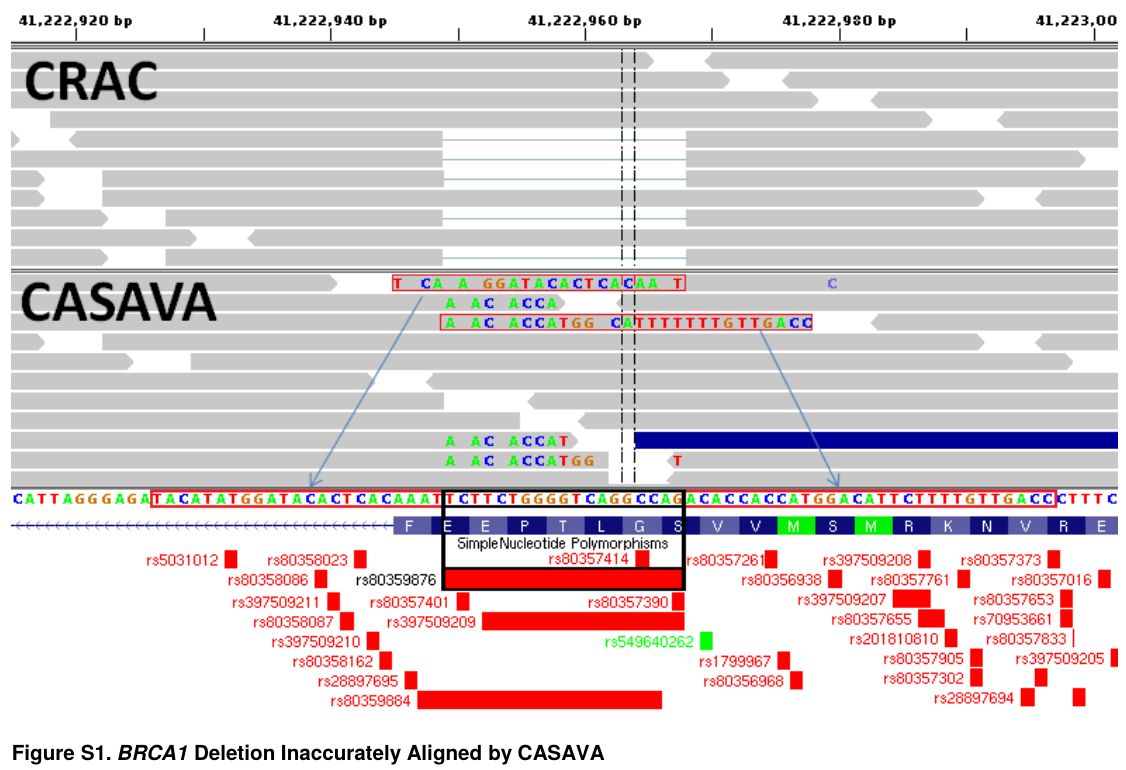
May 20, 2016Mucaki et al published an article in BMC Medical Genomics entitled “A unified analytic framework for prioritization of non-coding variants of uncertain significance in heritable breast and ovarian cancer”.
Their pipeline uses both CRAC and Casava for the sequence alignment, then SNVs and indels were called using GATK. There is little agreement between both methods and the authors exhibit an example of a discordance, which is at CRAC’s advantage.
Warning on libhts version
May 19, 2016Several users had the same issue, with libhts.
You may not have the good version of this library. You should install libhts1, which doesn’t seem to be directly available under Ubuntu 14.04.
You can find more details on that issue in the FAQ.
CRAC version 2.5.0 released
Jan 29, 2016Bug corrections
- Correct bug in chimeric alignment where alignments pos and chr where not properly setted
- Correct memory leak detected with valgrind
- Correct problem when second end has the length of the k-mer (Bug submitted by E. Mathijs)
- Correct segfault problem produced by a very special case of chimeric alignments (Bug submitted by E. Mathijs)
- Correct canonical k-mer counts from JellyReadsIndex when –stranded option is activated (Bug submitted by S. Beaumeunier)
CRAC version 2.4.0 released
Oct 2, 2015Improvements
- integrate a better crac_score for chimera events detection
- add a XS field as topHat does (it allows it to conform to cufflinks or stringtie)
Bug corrections
- extra fields are removed when the read is unmapped
- XE inconsistencies for SNVs and indels
- fixed wrong positions (reads and genome) for indels cases and add unit tests
CracTools version 1.2 released
Jul 17, 2015If you love CRAC and you want to get the most out of it, take a look at the new version of the CracTools that is a complete toolbox designed to build pipelines (in Perl) on top of CRAC.
It is also shipped with a command-line tool that extracts splicing variants, chimeric variants, indels and SNPs identified by CRAC at the read level then collapses them into standard formats (BED,VCF).
subscribe via RSS
News and updates
New releases and related tools will be announced through the CRAC mailing list.
Getting started
You want to try CRAC? Download it and check our getting started guide.
CracTools
Check out the CracTools : Integrated pipelines for RNA-Seq analysis using CRAC additional fields, annotation and much more.
Galaxy
A galaxy wrapper for CRAC is available and can be downloaded here.
Contact us
Any question regarding the software? Send an email to crac-bugs@lists.gforge.inria.fr.
A question, a comment, a remark on the algorithm or the article, send an email
to crac-article@lists.gforge.inria.fr
Publications
Nicolas Philippe, Mikaël Salson, Thérèse Commes and Eric Rivals.CRAC: an integrated approach to the analysis of RNA-seq reads doi:10.1186/gb-2013-14-3-r30